Get 1000 Hours free
On the UCSD Supercomputer
Start Your Trial
Movement of the A-strain maize streak virus in and out of Madagascar
Kehinde A. Oyeniran, Darren P. Martin, Jean-Michel Lett, Mbolarinosy Rasoafalimanana Rakotomalala, Hamza A. Azali, Arvind Varsani
Maize Streak Disease (MSD) is one of the most significant viral diseases of maize, the most important food crop in Africa. MSD’s impact on African maize crops results in at least $120M to $480M US in crop damage each year, with annual yield losses estimated at 6%–10% of total yield across the continent.
MSD is caused by a virus transmitted by leafhoppers of the genus Cicadulina (mainly C. mbila and C. storeyi). Several genetically distinct Maize Streak Virus (MSV) strains are known, and these can infect maize, sugarcane and a number of wild grasses in Southern Africa. MSV was the first plant virus described that contains circular single‐stranded DNA (ssDNA). MSV particles are twinned, quasi‐icosahedral (geminate), giving rise to the name ‘geminivirus’’.
MSV does not attack plants directly, rather transmission only occurs when leafhoppers ingest material from infected plants, and transmit the virus as they move on to feed on uninfected plants. Given its dependence on leafhoppers of genus Cicadulina for transmission, MSV is currently confined to regions of Africa where the genus Cicadulina occurs naturally. However, there is no obvious reason these vectors could not survive in climatically similar areas such as the southern USA and South America and Eurasia. If Cicadulina were to establish itself in a new region, maize crops outside of Africa could be seriously damaged. Accordingly, understanding how the virus spreads naturally within the African continent and across natural barriers is of significant interest.
With this in mind, Oyeniran et al. conducted a detailed study of the movements of MSV from mainland Africa to Madagascar and Comoros, nearby islands with no land bridge to the continent. Migration to these islands can only occur through relocation of infected leafhoppers by physical (human driven) transfer. To understand the how MSV spreads, the authors analyzed 524 complete MSV genome sequences collected from southern Africa, East Africa, and Indian Ocean islands between 1979 and 2010. The analysis included 56 MSV genome sequences from Madagascar (12 locations) and the Comoros (2 locations) collected between 2009 and 2010.
All specimens were found to be examples of the MSV-A variant, the most economically destructive of the MSV subtypes. The MSV-A virus has been categorized into different subtypes that describe the relationships between sequences and indicate the severity of symptoms caused by different groups of MSV-A isolates. All Madagascan isolates in the study belonged to the MSV-A1 subtype, belonging to one of three different known recombinant lineages: XI, IV, and V.
The authors analyzed their data using tools available through CIPRES, including MUSCLE for alignment, jModelTest2 for model selection, IQ-TREE to create maximum likelihood trees, and BEAST for subsequent Bayesian phylogeographic analyses. Their BEAST analyses used sampling year and sampling country metadata to inform strict molecular clock and discrete phylogeography models.
Discrete phylogeographic analysis suggested nine individual introductions of MSV-A from the continent to Madagascar between 1975 and 2003 (Figure 1), while a subsequent continuous analysis suggested perhaps five in the same time range. The Madagascan MSV-A viruses seemed to derive mostly from recombinant lineages initially identified in Uganda. Once established in Madagascar, MSV-A variants spread across the island at an average rate of roughly 47.6 km per year after separate introductions.
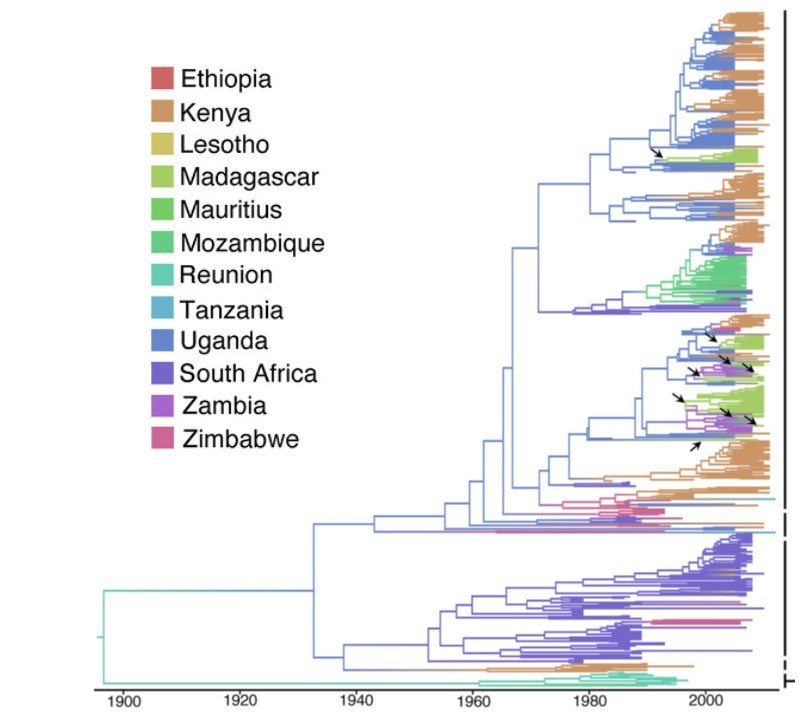
Figure 1. Discrete phylogeographic analysis of MSV isolates from Africa. The arrows note nine discrete introductions of MSV into Madagascar.
Takeaways:
